Before you begin working with Hyperchem, you should set some preferences in the program. First, click onSelect on the menu bar, then click on Multiple Selections. This option allows you to select more than one atom at a time. Next, click on Display on the menu bar, click on Labels… and click on the radio button next to Symbol under “Atoms”. This option turns on the display of the atomic symbol for each atom in the display window.
Drawing Bonds
Click on the Drawing Tool button. Click and drag in the display window. To draw a bond to an existing atom, click on the atom and drag.
Deleting Atoms or Bonds
Click on the Drawing Tool button. Right-click on any atom or bond to delete it.
Changing the Atom Type
Double-click on the Drawing Tool button (a Periodic Table will appear). Select the element by clicking on it in the Periodic Table. Close the Periodic Table window. Click on any atom in the display window to change the atom to the currently selected atom type.
Changing the Number of Bonds Between Two Atoms
Click on the Drawing Tool icon. Single, double, triple and resonance-type bonds can be selected by repeated clicking on the bond.
Model Building a Structure
Double-click on the Selection Tool button. Note that this will also add hydrogen atoms to any atoms that do not have a completely filled valence.
- Note: the “model build” operation will perform a “quick” optimization of the molecular geometry based on standard bond lengths and bond angles. You should always “model build” your structure before starting any higher level geometry optimizations (such as AM1).
Selecting Atoms
First, click on Select on the Toolbar (a menu will appear). If there is no check mark next to Multiple Selections, click on Multiple Selections. Then, click on the Selection Tool button. Click on the atoms to be selected (the selected atoms will become highlighted). To select all atoms in the display window, double-click in any blank area of the workspace.
- Note: you must “deselect” any selected atoms before starting a geometry optimization.
Deselecting Atoms
Click on the Selection Tool button. Right-click on the atoms to be deselected. To deselect all selected atoms, right-click in any blank area of the workspace.
Rotating a Molecule
Click on either the XY Rotation Tool or the Z Rotation Tool button. Click and drag in the workspace to rotate the molecule.
Moving a Molecule
Click on the XY Translation Tool button. Click and drag in the workspace to move the molecule.
Resizing
Click on the Zoom Tool button. Click and drag in the workspace to resize the molecule.
Changing the Display Type
Click on Display on the menu bar (a menu will appear). Click on Rendering… Select the type of display for the molecule.
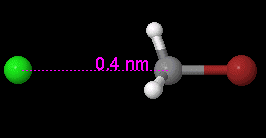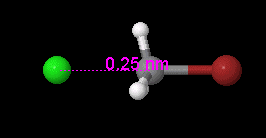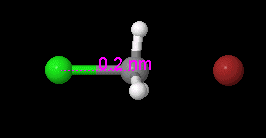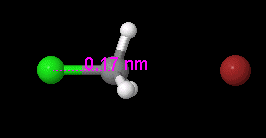
Setting a Bond Length
Click on the Selection Tool button. Click on the two atoms which are bonded (the two atoms and the bond between them will become highlighted). Click on Edit on the menu bar. Click on Set Bond Length…, and enter a value (in Angstroms) for the bond length. Click OK.
Setting a Bond Angle
Click on the Selection Tool button. Click on the three atoms which define the bond angle (the three atoms and the bonds between them will become highlighted). Click on Edit on the menu bar. Click on Set Bond Angle…, and enter a value (in degrees) for the bond angle. Click OK.
Setting a Bond Torsion Angle
Click on the Selection Tool button. Click on the four atoms which define the bond torsion angle (the four atoms and the bonds between them will become highlighted). Click on Edit on the menu bar. Click on Set Bond Torsion…, and enter a value (in degrees) for the bond torsion angle. Click OK.
Optimizing the Geometry of a Molecule Using the Molecular Mechanics Method (MM+)
Click on Setup on the menu bar. Click on Molecular Mechanics… Under “Method”, click on the radio button next to “MM+”. Click OK.
Click on Compute on the menu bar. Click on Geometry Optimization… Click on OK.
Optimizing the Geometry of a Molecule Using a Semi-Empirical Method
Click on Setup on the menu bar. Click on Semi-empirical… Under “Methods”, click on the radio button next to the method to be used (e.g., “AM1″). Click on the “Options…” button. Under “Charge and Spin”, enter the “Total charge” of the molecule or ion. Click OK. Click OK.
Click on Compute on the menu bar. Click on Geometry Optimization… Click on OK.
Calculating the Energy of a Molecule For a Specific Geometry Using a Semi-Empirical Method (Single Point)
Click on Setup on the menu bar. Click on Semi-empirical… Under “Methods”, click on the radio button next to the method to be used (e.g., “AM1″). Click on the “Options…” button. Under “Charge and Spin”, enter the “Total charge” of the molecule or ion. Click OK. Click OK.
Click on Compute on the menu bar. Click on Single Point. Click on OK.
Calculating the Orbitals of a Molecule Using a Semi-Empirical Method
Click on Compute on the menu bar. Click on Orbitals… In the orbital energy diagram, click on the orbital to be plotted (the line representing the orbital will be highlighted). Click on “Plot”.
- Note: you must perform a “single point” calculation before you can calculate the orbitals of the molecule.
Displaying the Values for Bond Lengths, Bond Angles and Bond Torsion Angles
Click on the Selection Tool button. Click on the two, three or four atoms which define the bond length, bond angle or bond torsion angle, respectively. The value (in Angstroms or degrees) will be displayed on the Status Line.
Calculating the Vibrational Spectrum of a Molecule Using a Semi-Empirical Method
Click on Compute on the menu bar. Click on Vibrations. When the calculation has finished, click onCompute on the menu bar again and then click on Vibrational Spectrum… Click on any of the lines at the top of the “Vibrational Spectrum” window (the line will become highlighted) to display the frequency of that vibration.
To animate a particular vibration, first make sure there is a check mark next to Animate vibrations in the “Vibrational Spectrum” window (if there is not, click on the check box). Then, click on the line at the top of the “Vibrational Spectrum” window which represents the vibration that you want to animate (the line will become highlighted). Click OK. Click Cancel on the menu bar to stop the animation.
Source: kasmui.blog.com